18.10 Post MICE data management
Sometimes you’ll have a need to do additional data management after imputation has been completed. Creating binary indicators of an event, re-creating scale variables etc. The general approach is to transform the imputed data into long format using complete with the argument include=TRUE , do the necessary data management, and then convert it back to a mids object type.
Continuing with the penguin example, let’s create a new variable that is the ratio of bill length to depth.
Recapping prior steps of imputing, and then creating the completed long data set.
## imp_pen <- mice(pen.miss, m=10, maxit=25, meth="pmm", seed=500, printFlag=FALSE)
pen_long <- complete(imp_pen, 'long', include=TRUE)We create the new ratio variable on the long data:
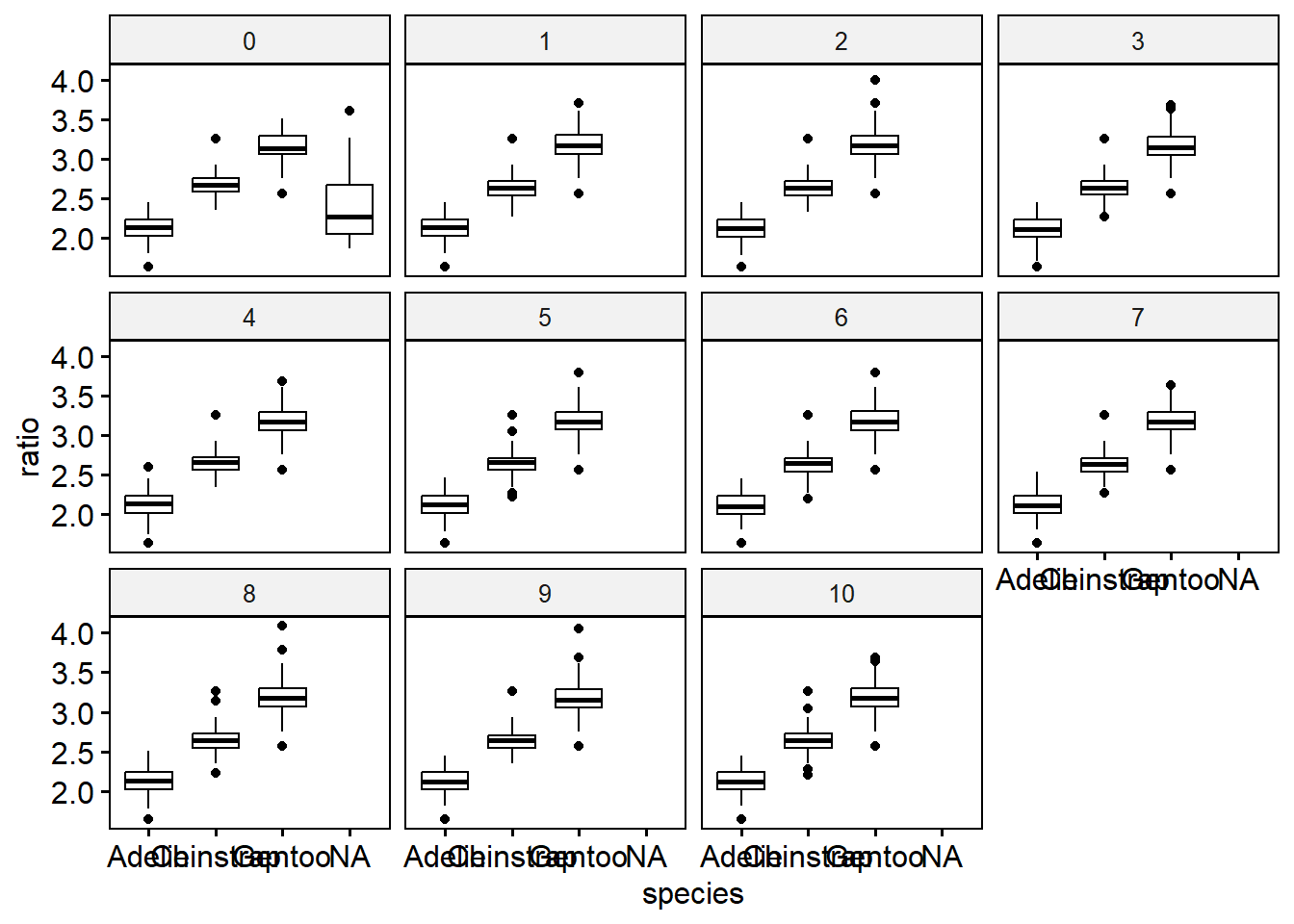
Let’s visualize this to see how different the distributions are across imputation. Notice imputation “0” still has missing data - this is a result of using include = TRUE and keeping the original data as part of the pen_long data.
Then convert the data back to mids object, specifying the variable name that identifies the imputation number.
Now we can conduct analyses such as an ANOVA (in linear model form) to see if this ratio differs significantly across the species.
nova.ratio <- with(imp_pen1, lm(ratio ~ species))
pool(nova.ratio) |> summary()
## term estimate std.error statistic df p.value
## 1 (Intercept) 2.1221949 0.01435687 147.81738 281.7994 4.610006e-269
## 2 speciesChinstrap 0.5217842 0.02569344 20.30807 245.0315 1.958440e-54
## 3 speciesGentoo 1.0643029 0.02165568 49.14659 254.5258 6.526634e-132