18.1 Identifying missing data
- Missing data in
Ris denoted asNA - Arithmetic functions on missing data will return missing
survey <- MASS::survey # to avoid loading the MASS library which will conflict with dplyr
head(survey$Pulse)
## [1] 92 104 87 NA 35 64
mean(survey$Pulse)
## [1] NAThe summary() function will always show missing.
summary(survey$Pulse)
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## 35.00 66.00 72.50 74.15 80.00 104.00 45The is.na() function is helpful to identify rows with missing data
The function table() will not show NA by default.
table(survey$M.I)
##
## Imperial Metric
## 68 141
table(survey$M.I, useNA="always")
##
## Imperial Metric <NA>
## 68 141 28- What percent of the data set is missing?
4% of the data points are missing.
- How much missing is there per variable?
prop.miss <- apply(survey, 2, function(x) round(sum(is.na(x))/NROW(x),4))
prop.miss
## Sex Wr.Hnd NW.Hnd W.Hnd Fold Pulse Clap Exer Smoke Height M.I
## 0.0042 0.0042 0.0042 0.0042 0.0000 0.1899 0.0042 0.0000 0.0042 0.1181 0.1181
## Age
## 0.0000The amount of missing data per variable varies from 0 to 19%.
18.1.1 Visualize missing patterns
Using ggplot2
pmpv <- data.frame(variable = names(survey), pct.miss =prop.miss)
ggplot(pmpv, aes(x=variable, y=pct.miss)) +
geom_bar(stat="identity") + ylab("Percent") + scale_y_continuous(labels=scales::percent, limits=c(0,1)) +
geom_text(data=pmpv, aes(label=paste0(round(pct.miss*100,1),"%"), y=pct.miss+.025), size=4)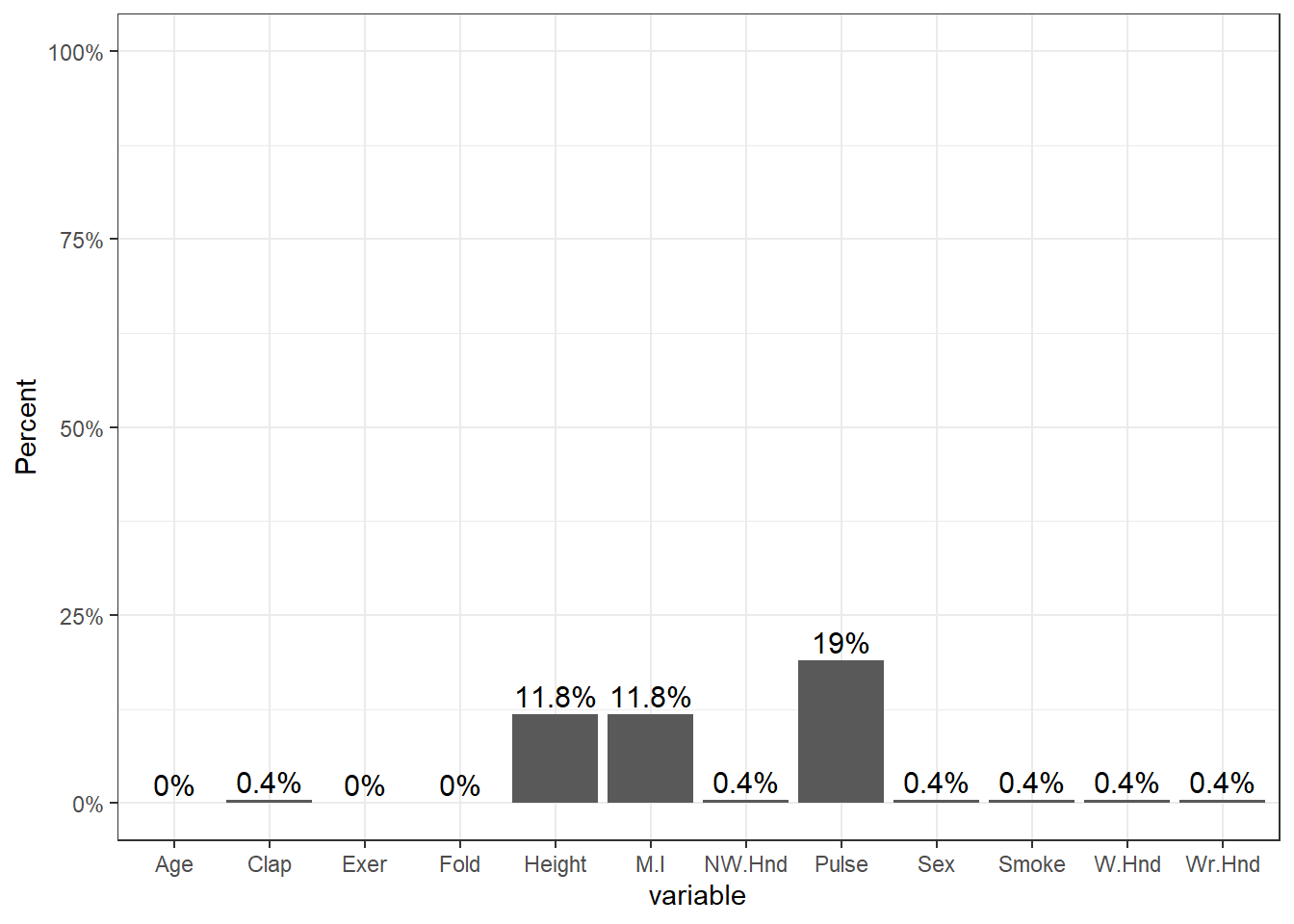
Using mice
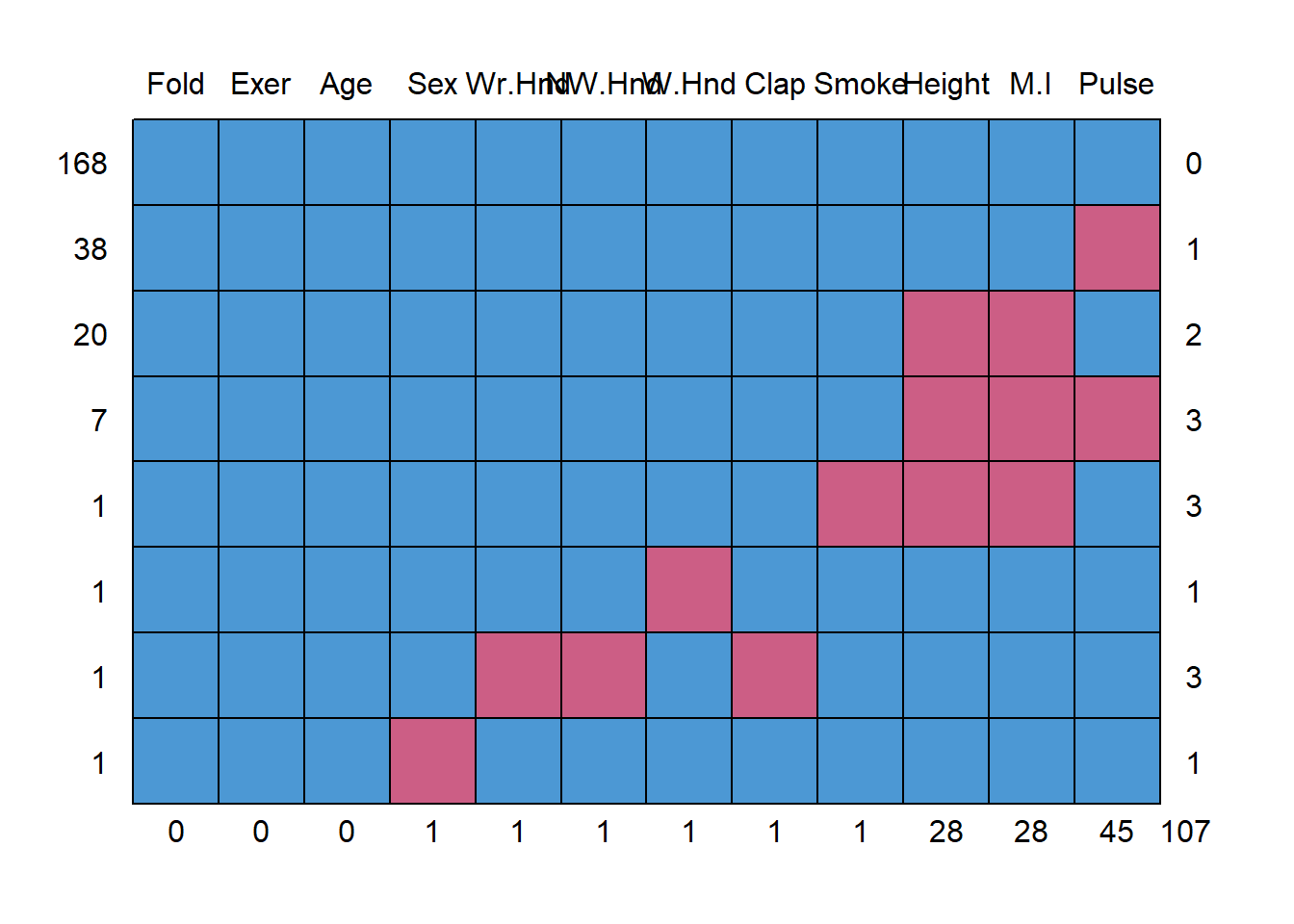
## Fold Exer Age Sex Wr.Hnd NW.Hnd W.Hnd Clap Smoke Height M.I Pulse
## 168 1 1 1 1 1 1 1 1 1 1 1 1 0
## 38 1 1 1 1 1 1 1 1 1 1 1 0 1
## 20 1 1 1 1 1 1 1 1 1 0 0 1 2
## 7 1 1 1 1 1 1 1 1 1 0 0 0 3
## 1 1 1 1 1 1 1 1 1 0 0 0 1 3
## 1 1 1 1 1 1 1 0 1 1 1 1 1 1
## 1 1 1 1 1 0 0 1 0 1 1 1 1 3
## 1 1 1 1 0 1 1 1 1 1 1 1 1 1
## 0 0 0 1 1 1 1 1 1 28 28 45 107This somewhat ugly output tells us that 168 records have no missing data, 38 records are missing only Pulse and 20 are missing both Height and M.I.
Using VIM
library(VIM)
aggr(survey, col=c('chartreuse3','mediumvioletred'),
numbers=TRUE, sortVars=TRUE,
labels=names(survey), cex.axis=.7,
gap=3, ylab=c("Missing data","Pattern"))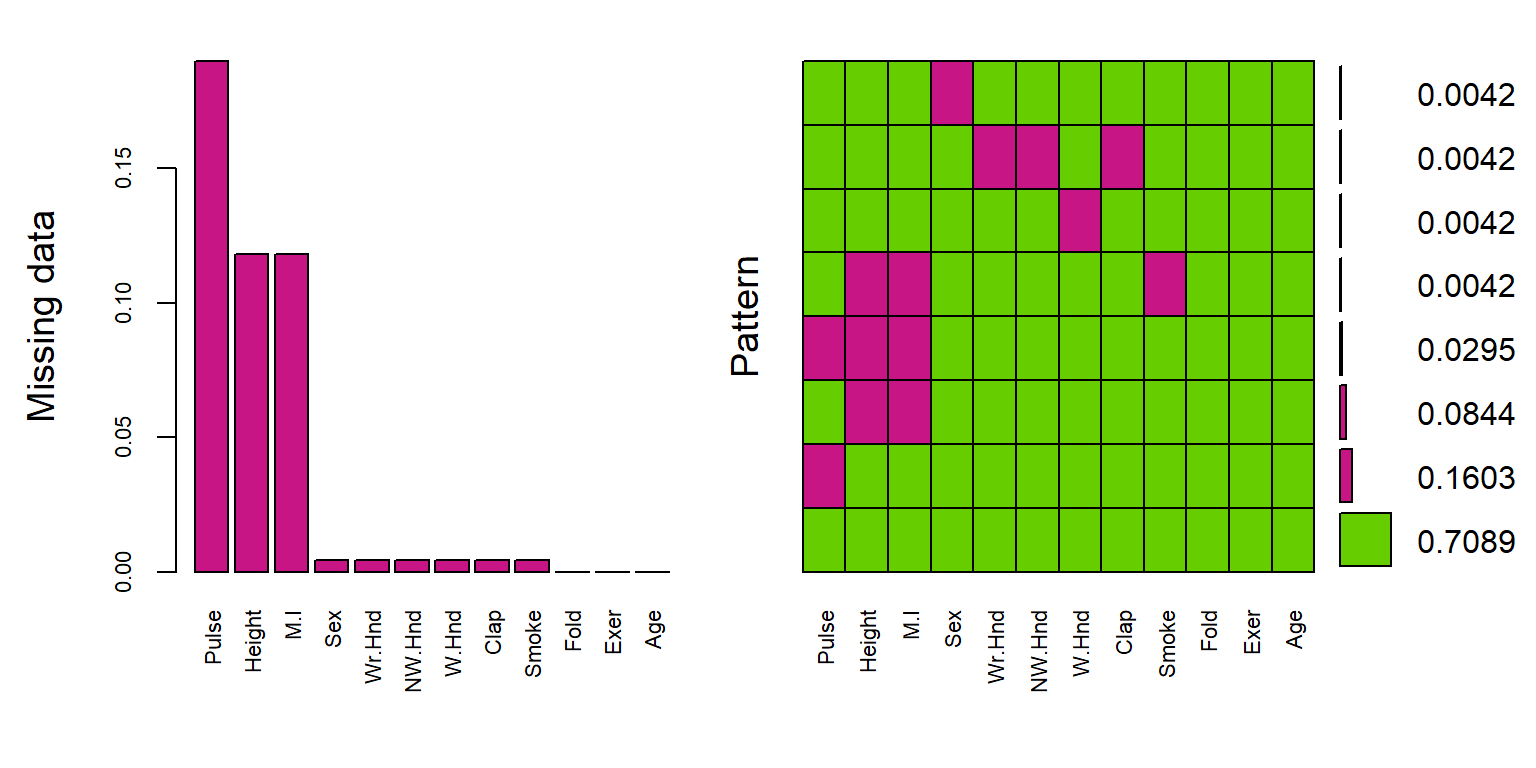
The plot on the left is a simplified, and ordered version of the ggplot from above, except the bars appear to be inflated because the y-axis goes up to 15% instead of 100%.
The plot on the right shows the missing data patterns, and indicate that 71% of the records has complete cases, and that everyone who is missing M.I. is also missing Height.
Another plot that can be helpful to identify patterns of missing data is a marginplot (also from VIM).
- Two continuous variables are plotted against each other.
- Blue bivariate scatterplot and univariate boxplots are for the observations where values on both variables are observed.
- Red univariate dotplots and boxplots are drawn for the data that is only observed on one of the two variables in question.
- The darkred text indicates how many records are missing on both.
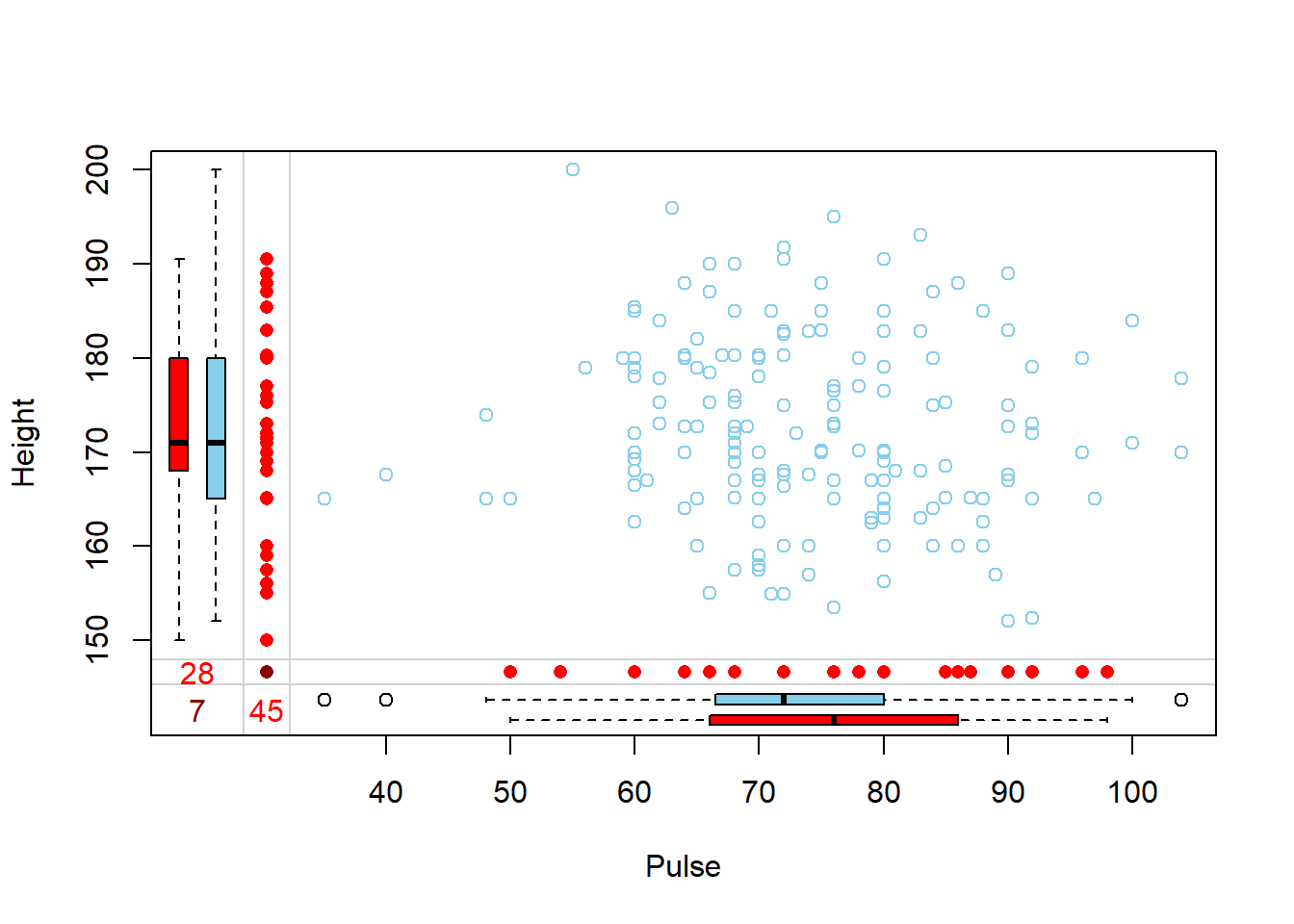
This shows us that the observations missing pulse have the same median height, but those missing height have a higher median pulse rate.
18.1.2 Example: Parental HIV
18.1.2.2 Examine missing data patterns of scale variables.
The parental bonding and BSI scale variables are aggregated variables, meaning they are sums or means of a handful of component variables. That means if any one component variable is missing, the entire scale is missing. E.g. if y = x1+x2+x3, then y is missing if any of x1, x2 or x3 are missing.
scale.vars <- hiv %>% select(parent_care:bsi_psycho, gender, siblings, age)
aggr(scale.vars, sortVars=TRUE, combined=TRUE, numbers=TRUE, cex.axis=.7)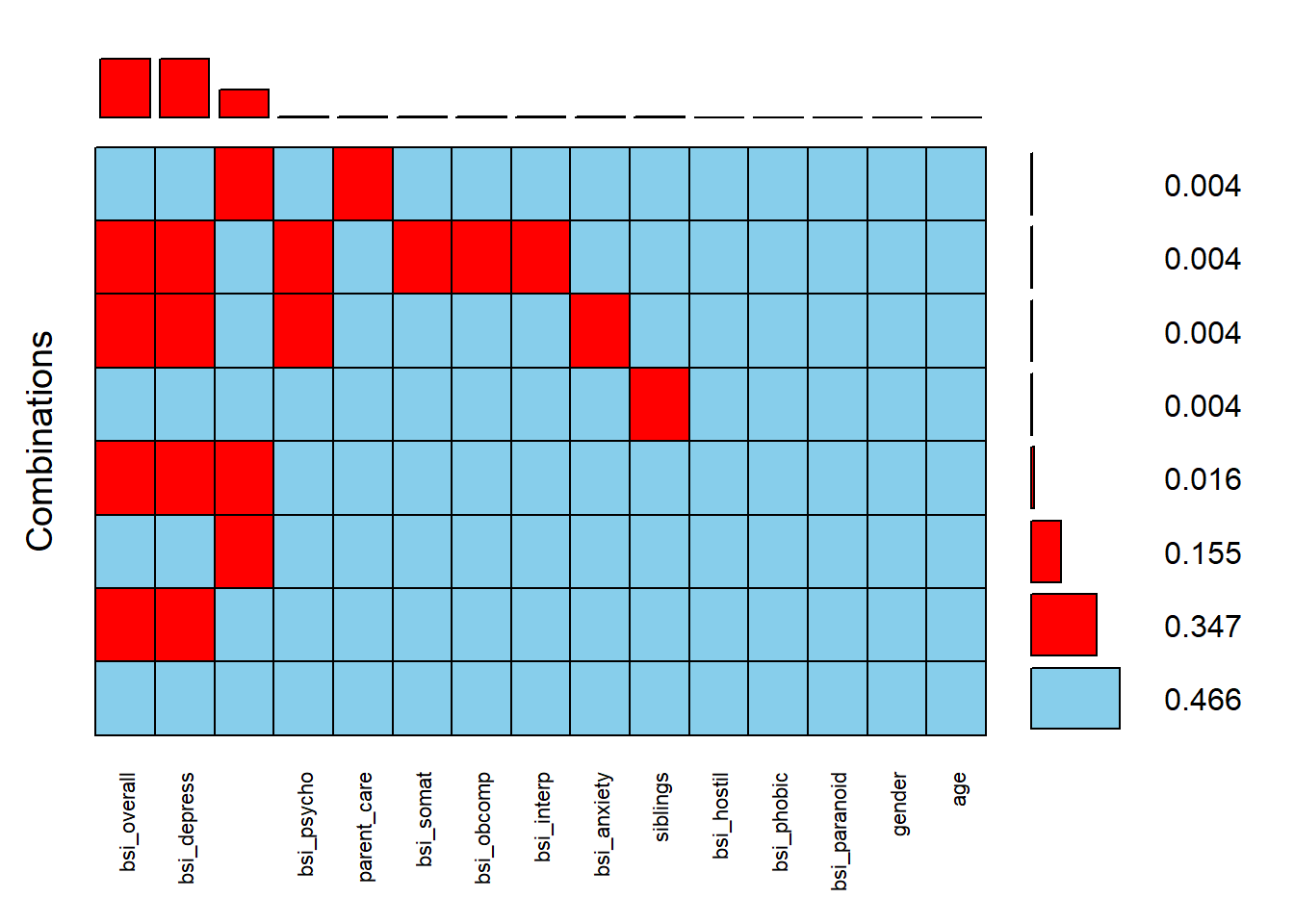
##
## Variables sorted by number of missings:
## Variable Count
## bsi_overall 93
## bsi_depress 93
## parent_overprotection 44
## bsi_psycho 2
## parent_care 1
## bsi_somat 1
## bsi_obcomp 1
## bsi_interp 1
## bsi_anxiety 1
## siblings 1
## bsi_hostil 0
## bsi_phobic 0
## bsi_paranoid 0
## gender 0
## age 034.7% of records are missing both bsi_overall and bsi_depress This makes sense since bsi_depress is a subscale containing 9 component variables and the bsi_overall is an average of all 52.
Another 15.5% of records are missing parental_overprotection.
Is there a bivariate pattern between missing and observed values of bsi_depress and parent_overprotection?
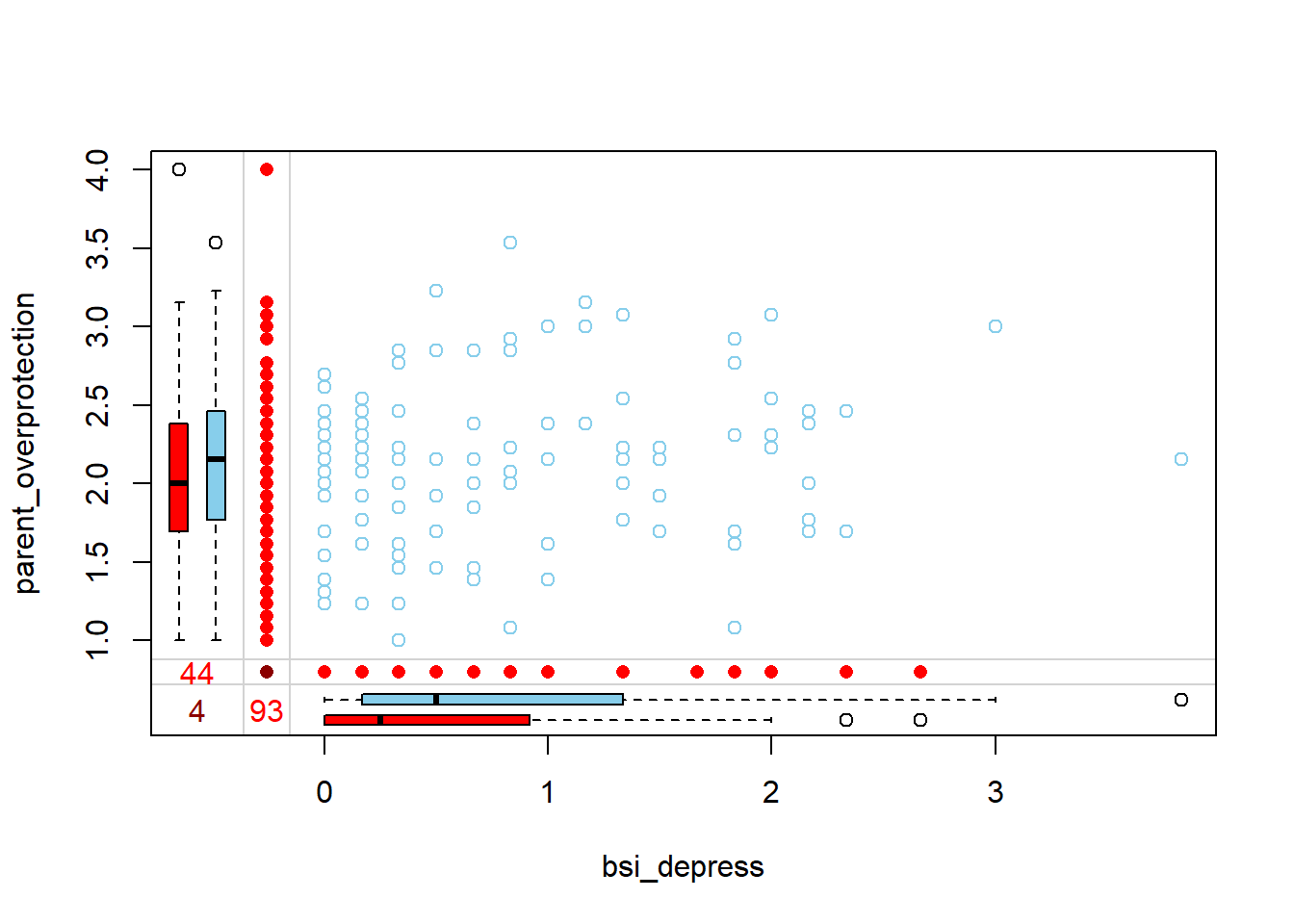
When someone is missing parent_overprotection, they have a lower bsi_depress score. Those missing bsi_depress have a slightly lower parental_overprotection score. Only 4 individuals are missing both values.