14.4 Generating PC’s using R
Corresponding reading: PMA6 Ch 14.3-14.4
Calculating the principal components in R can be done using the function prcomp() and princomp(). This section of notes uses princomp(), not for any specific reason. STHDA has a good overview of the difference between prcomp() and princomp(). It appears that prcomp() may have some more post-analysis fancy visualizations available.
- Requirements of
data: This must be a numeric matrix. Since I made up this example by generating data in section 14.2, I know they are numeric.
pr <- princomp(data)
summary(pr)
## Importance of components:
## Comp.1 Comp.2
## Standard deviation 11.4019265 4.2236767
## Proportion of Variance 0.8793355 0.1206645
## Cumulative Proportion 0.8793355 1.0000000- The summary output above shows the first PC (
Comp.1) explains the highest proportion of variance. - The values for the matrix \(\mathbf{A}\) is contained in
pr$loadings.
pr$loadings
##
## Loadings:
## Comp.1 Comp.2
## X1 0.854 0.519
## X2 0.519 -0.854
##
## Comp.1 Comp.2
## SS loadings 1.0 1.0
## Proportion Var 0.5 0.5
## Cumulative Var 0.5 1.0To visualize these new axes, we plot the centered data.
a <- pr$loadings
x1 <- with(data, X1 - mean(X1))
x2 <- with(data, X2 - mean(X2))
plot(c(-40, 40), c(-20, 20), type="n",xlab="x1", ylab="x2")
points(x=x1, y=x2, pch=16)
abline(0, a[2,1]/a[1,1]); text(30, 10, expression(C[1]))
abline(0, a[2,2]/a[1,2]); text(-10, 20, expression(C[2]))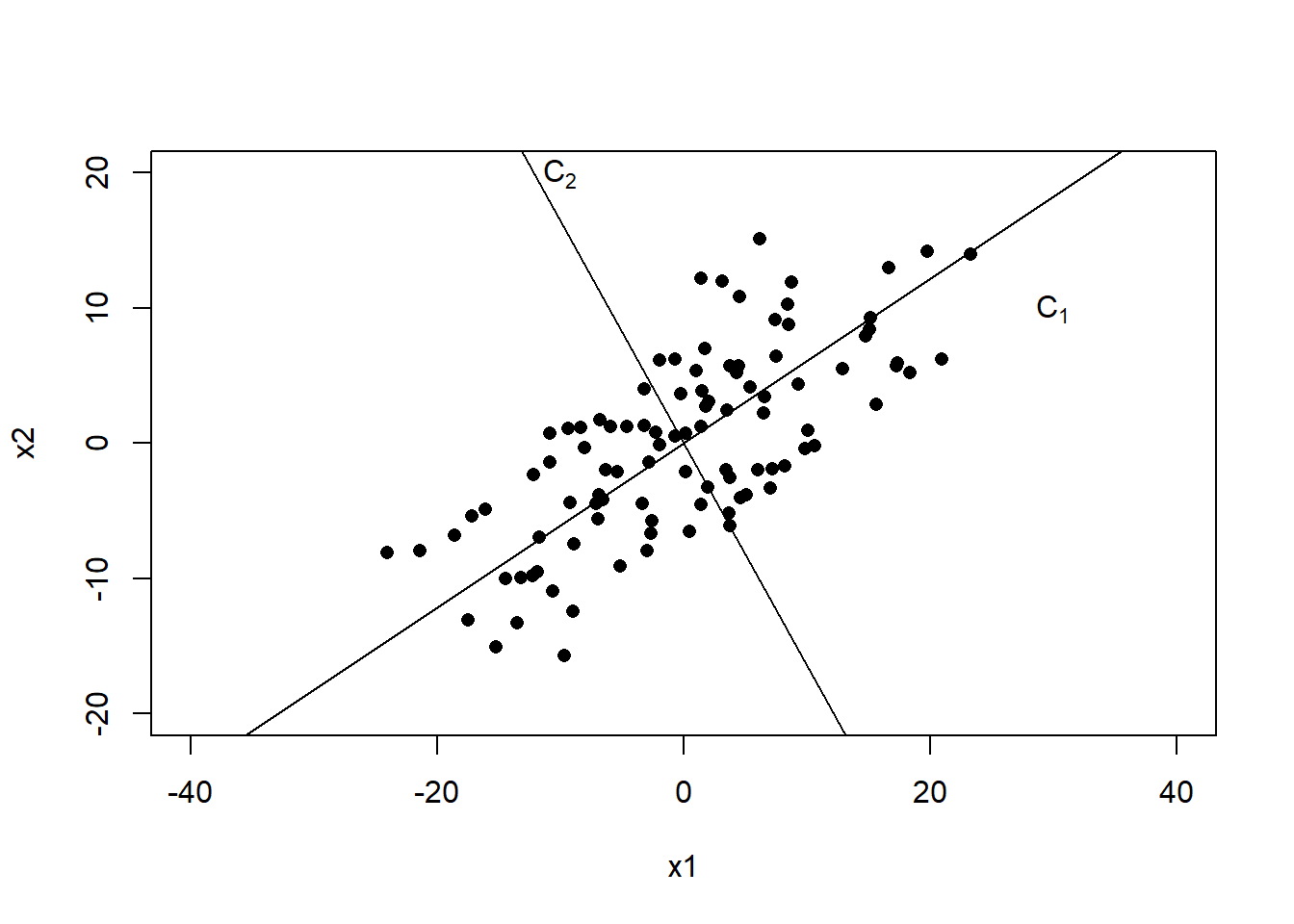
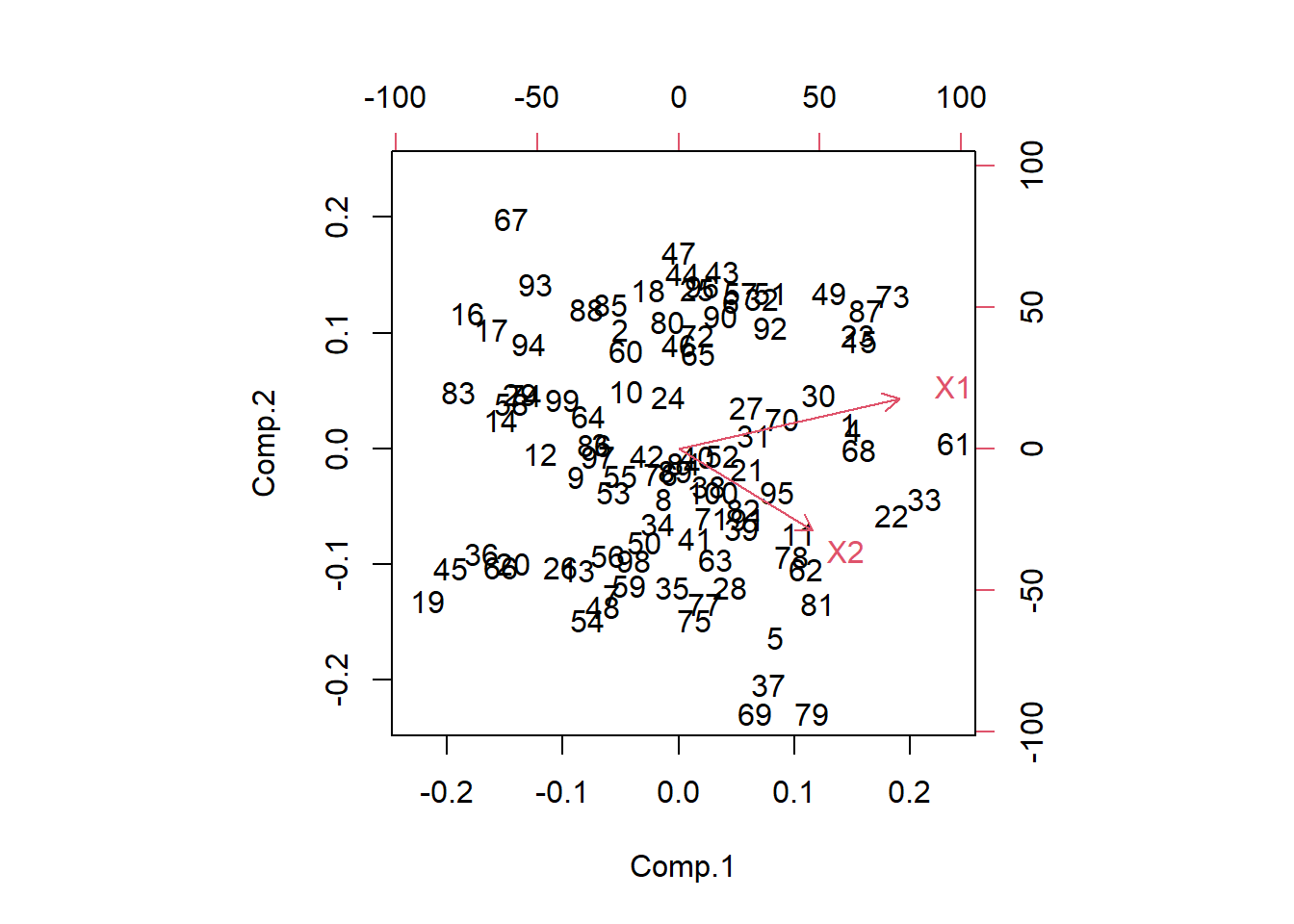
Plot the original data on the new axes we see that PC1 and PC2 are uncorrelated. The red vectors show you where the original coordinates were at.
biplot(pr)